MedeA MLP
At-a-Glance
MedeA®[1] MLP (Machine-Learned Potential) delivers comprehensive support for LAMMPS-based machine-learned potential simulations within the MedeA Environment. Simulate mechanical, vibrational, and transport properties with unprecedented efficiency, backed by the powerful analysis capabilities of the MedeA Environment.
MedeA MLP now features the GRACE 1L/2L Foundational models (OMAT & OAM) [2], representing a major advancement in materials modeling. These state-of-the-art models join an extensive library of published MLPs spanning diverse formalisms including the Spectral Neighbor Analysis Potential (SNAP) [3], Neural Network Potentials (NNP) [4], and the Atomic Cluster Expansion (ACE) [5], and the GRaph Atomic Cluster Expansion (GRACE) formalism [6], all fully supported by MedeA LAMMPS.
Key Benefits
Productivity
- Extends ab initio accuracy to dramatically larger length and time scales
- Streamlines workflow through intelligent automation and data management
Access
- Provides a collection of MLPs ready for use with MedeA LAMMPS simulations
- Gives access to the highly accurate and efficient foundational GRACE potentials
- Integrates with the MedeA MLPG (MedeA Machine-Learned Potential Generator) for creating custom potentials tailored to your research
- Enables LAMMPS simulations using MLPs generated from VASP MLFF calculations
Bridging Quantum Accuracy and Classical Scale
Machine Learning (ML) is revolutionizing materials science by enabling researchers to extract maximum value from rich first-principles datasets, allowing to achieve the accuracy and unbiased nature of ab initio methods while simulating realistic system sizes and timescales.
In particular, ML methods for energy and force calculation have been used for a number of years. In a remarkable paper published in 1998, Gassner et al. [7] demonstrated that “the advantages of a neural network type potential function as a model-independent and semiautomatic potential function outweigh the disadvantages in computing speed and lack of interpretability”. In recent years it has become increasingly clear that such approaches, employing novel descriptors and advanced ML techniques, can yield exceptionally accurate reproduction of quantum mechanical training data at substantially reduced computational cost [8].
Foundational Models: A New Era
The inclusion of GRACE 1L/2L foundational models marks a transformative moment for materials simulation [2]. These universal models, trained on vast and diverse datasets (including 110 million structures and covering 89 chemical elements with a focus on inorganic bulk materials [9]), provide exceptional transferability across chemical space, enabling accurate predictions for materials and compositions far beyond classical potential limitations.
The foundational GRACE potentials rank at the top of the MatBench Discovery leaderboard (https://matbench-discovery.materialsproject.org/) and establish a new ‘Pareto front’, offering the best available balance between high accuracy and computational speed (see Fig. 1).
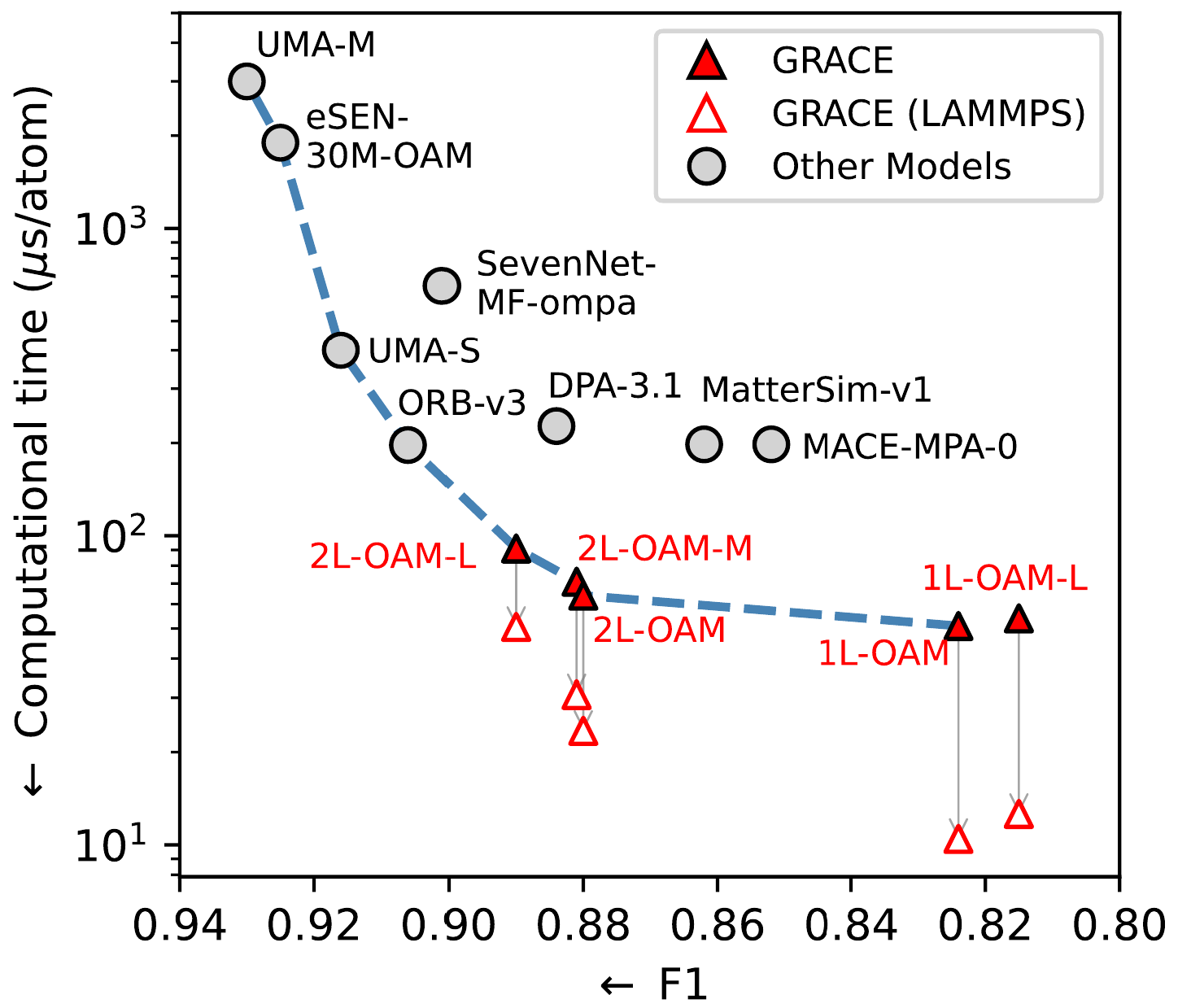
Model performance for stable structure identification (F1 score in MatBench Discovery benchmark) versus computational time per atom. A higher F1 score indicates better performance. The blue dashed line links Pareto optimal models (from Ref. 2).
MedeA MLP makes these cutting-edge methods effortlessly accessible. By bringing MLPs into the MedeA workflows, MedeA MLP extends ab initio precision to previously unreachable scales, while the intuitive MedeA Environment infrastructure ensures all input data and files are readily accessible.
Technical Features
User Interface
- Simple import of MLP .frc files
- Streamlined LAMMPS input generation
- Efficient preparation and deployment of simulations in the MedeA Environment
- Full integration with MedeA Builders and Property Modules for seamless system construction as well as streamlined property prediction
Example Systems
- Element-specific MLPs: Cu, Ge, Li, Mo, Ni, Si, Ta, W
- Compound-specific MLPs: InP, NbMoTaW, NiMo, WBe
- Foundational GRACE models: Broad coverage across the periodic table
Required Modules
- MedeA Environment
- MedeA LAMMPS
Find Out More
Learn more about Machine Learning by watching the webinars:
- https://www.materialsdesign.com/webinars/recorded/mlp-surpassing-the-limits-of-ab-initio
- https://www.materialsdesign.com/webinars/recorded/precision-at-scale-with-machine-learned-potentials
| [1] | MedeA and Materials Design are registered trademarks of Materials Design, Inc. |
| [2] | (1, 2) Y. Lysogorskiy et al., arXiv:2508.17936 (2025) (DOI) |
| [3] | A. P. Thompson et al., J. Comp. Phys. 285, 316 (2015) (DOI) |
| [4] | A. Singraber et al., (DOI) |
| [5] | R. Drautz, Phys. Rev. B 99, 014104 (2019) (DOI), Phys. Rev. B 100, 249901(E) (2019) (DOI) |
| [6] | A. Bochkarev et al., Phys. Rev. X 14, 021036 (2024) (DOI) |
| [7] | H. Gassner et al.,, J. Phys. Chem. A 102, 4596 (1998) (DOI) |
| [8] | V. Eyert et al., J. Mater. Res. 38, 5079 (2023) (DOI) |
| [9] | L. Barroso-Luque et al., arXiv:2410.12771 (2024) (DOI) |
| download: | pdf |
|---|